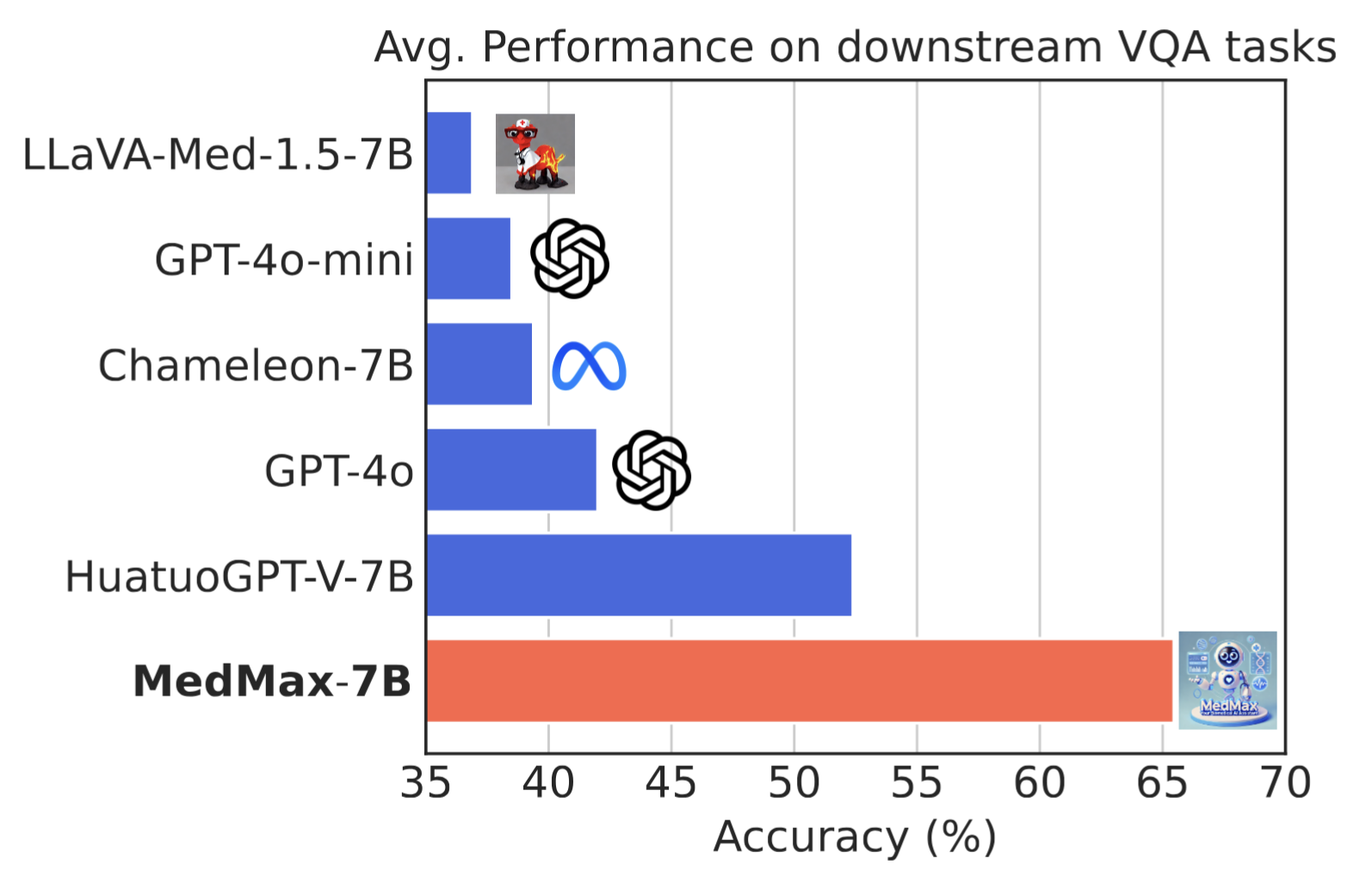
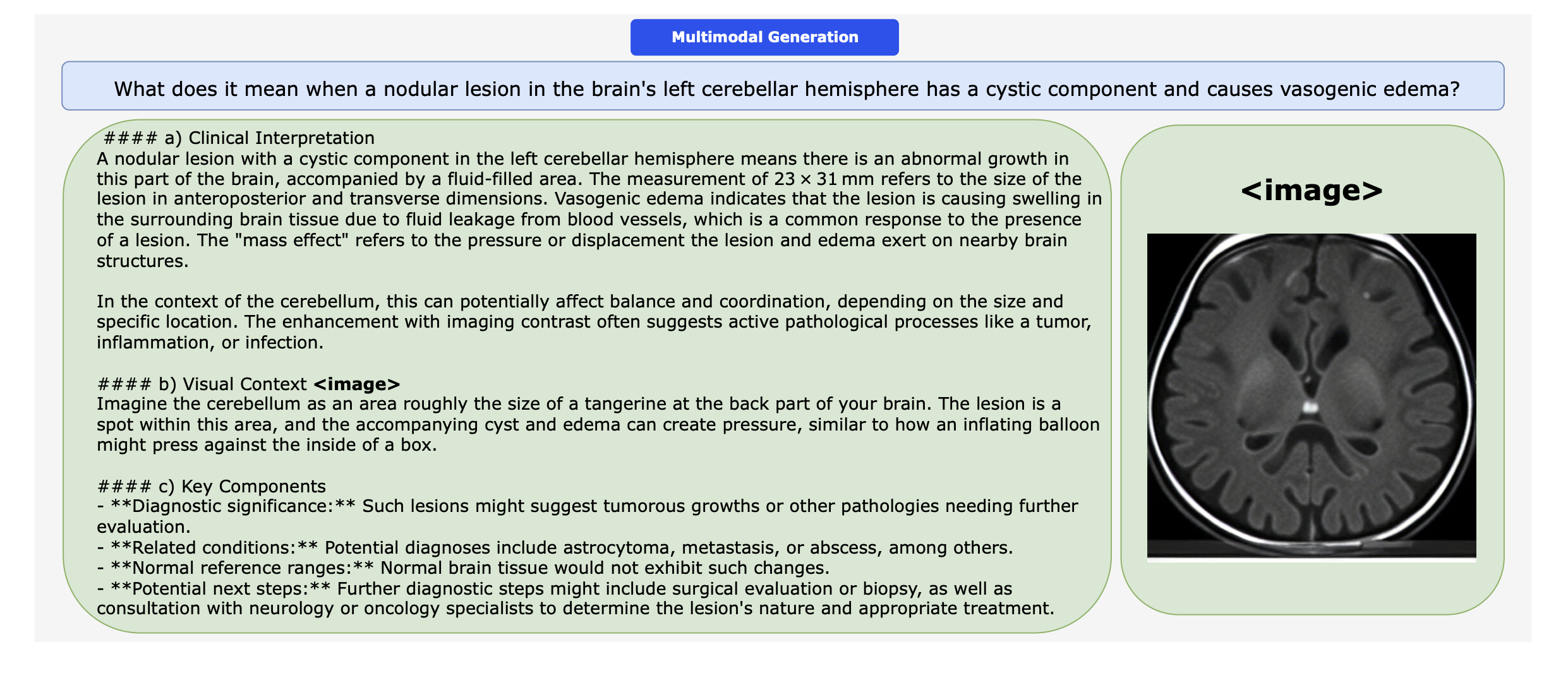
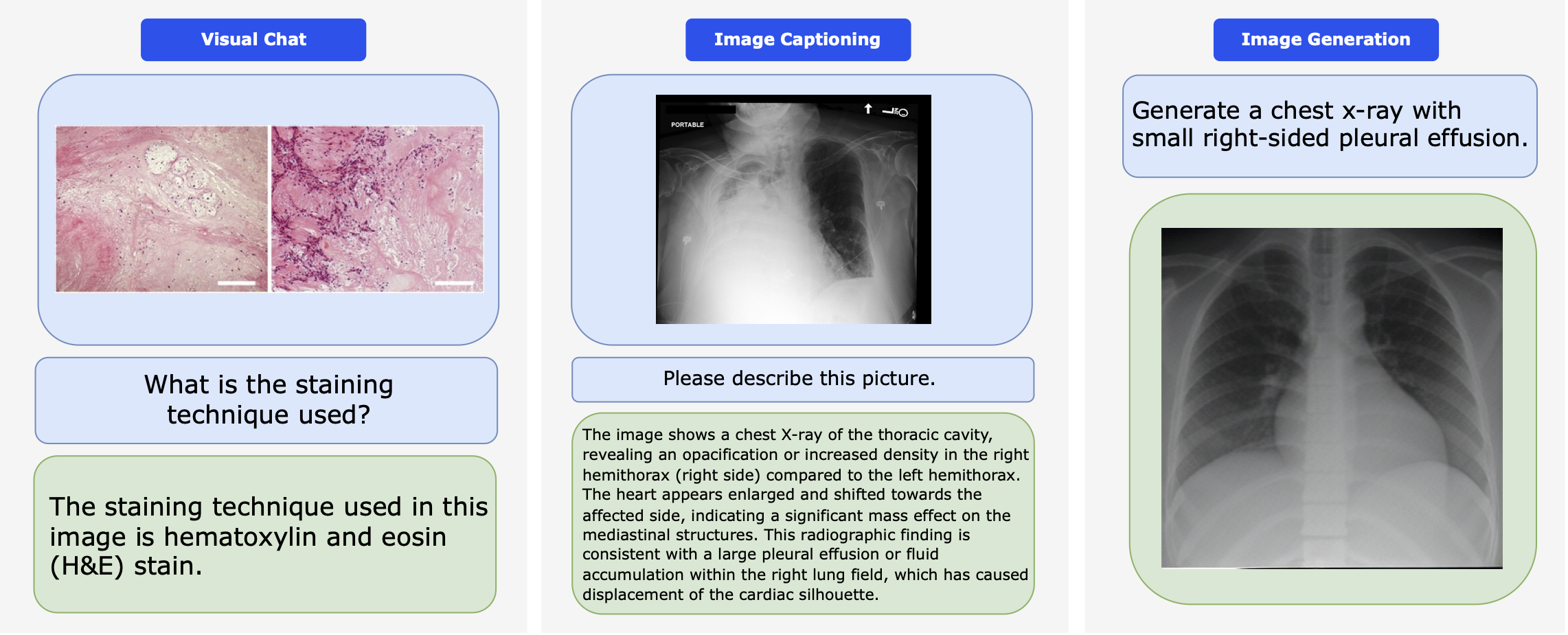
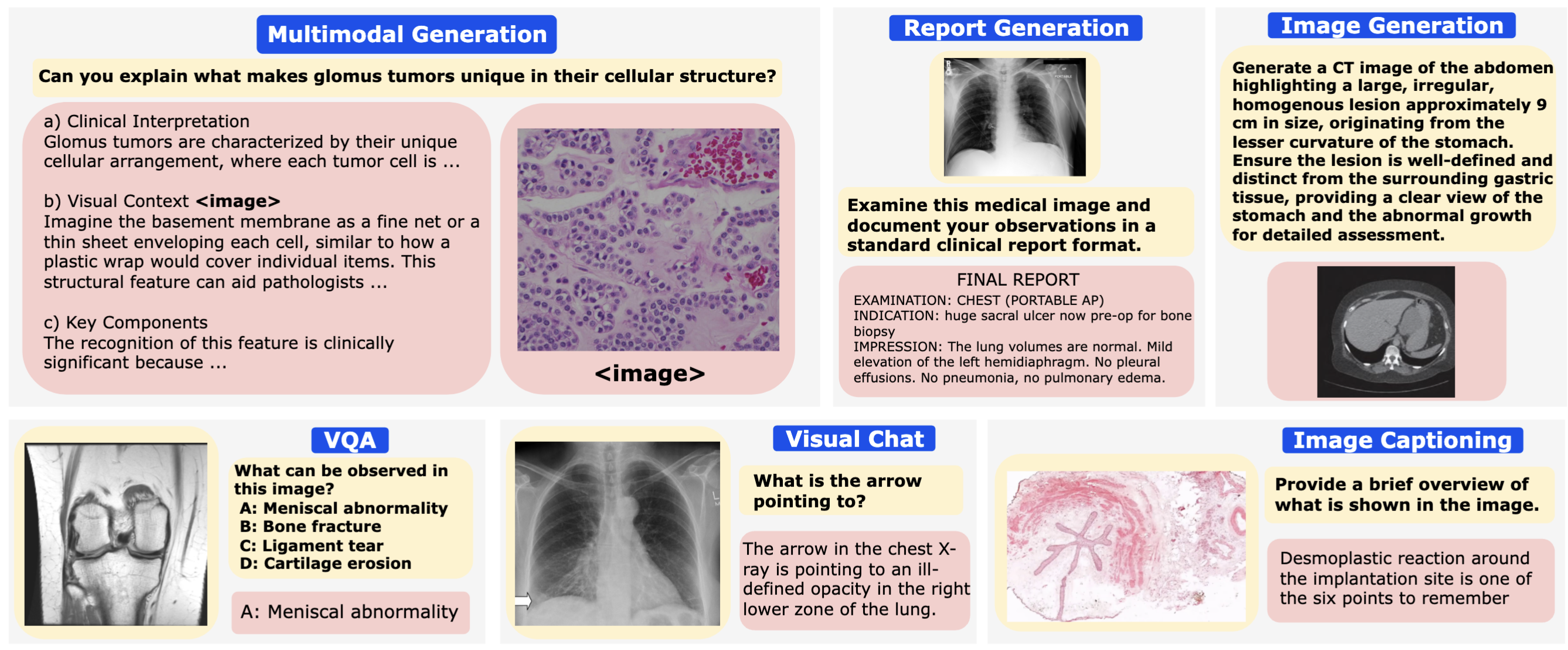
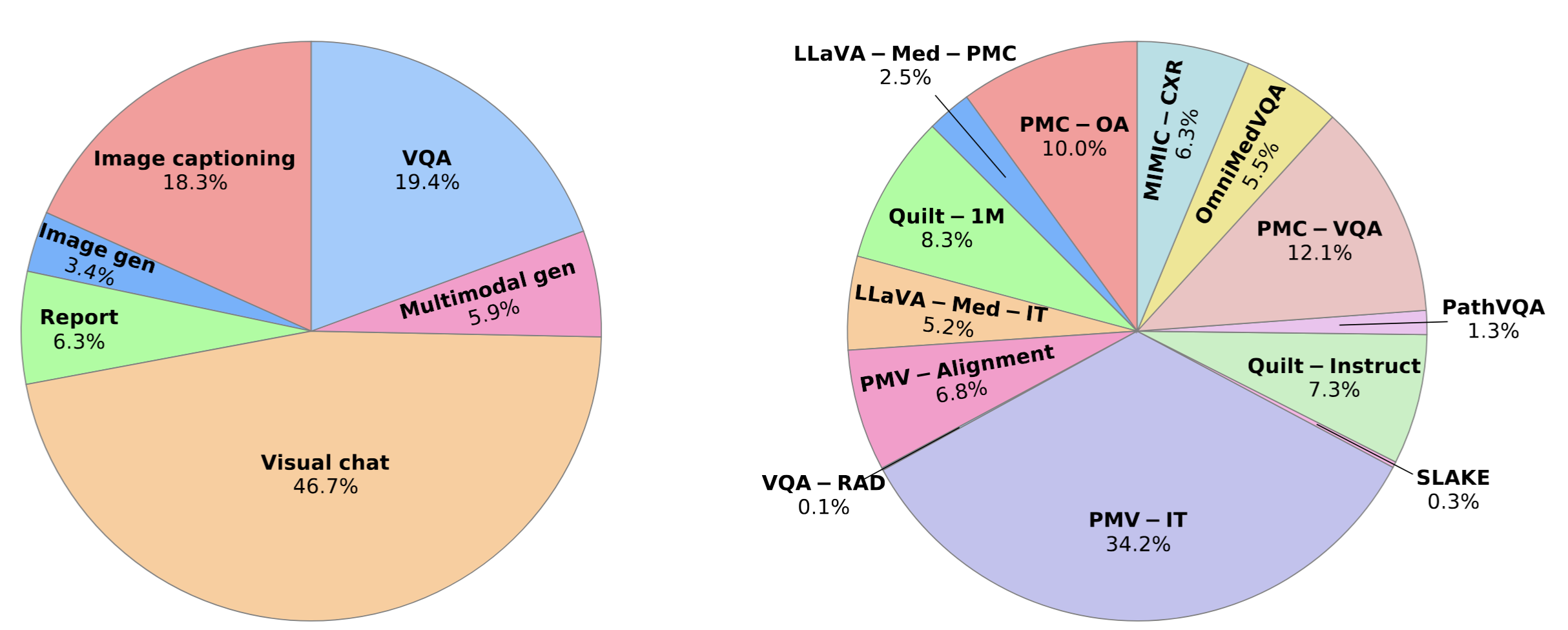
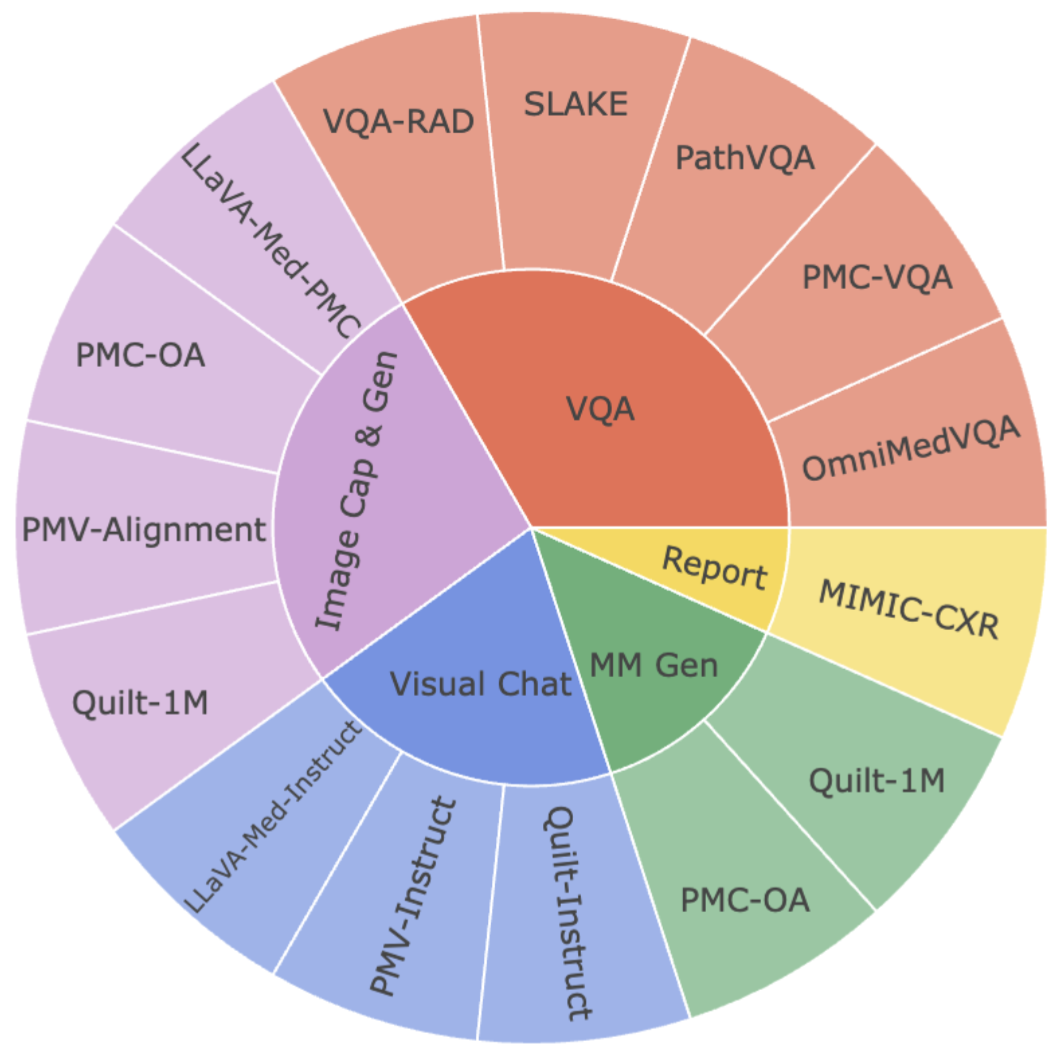
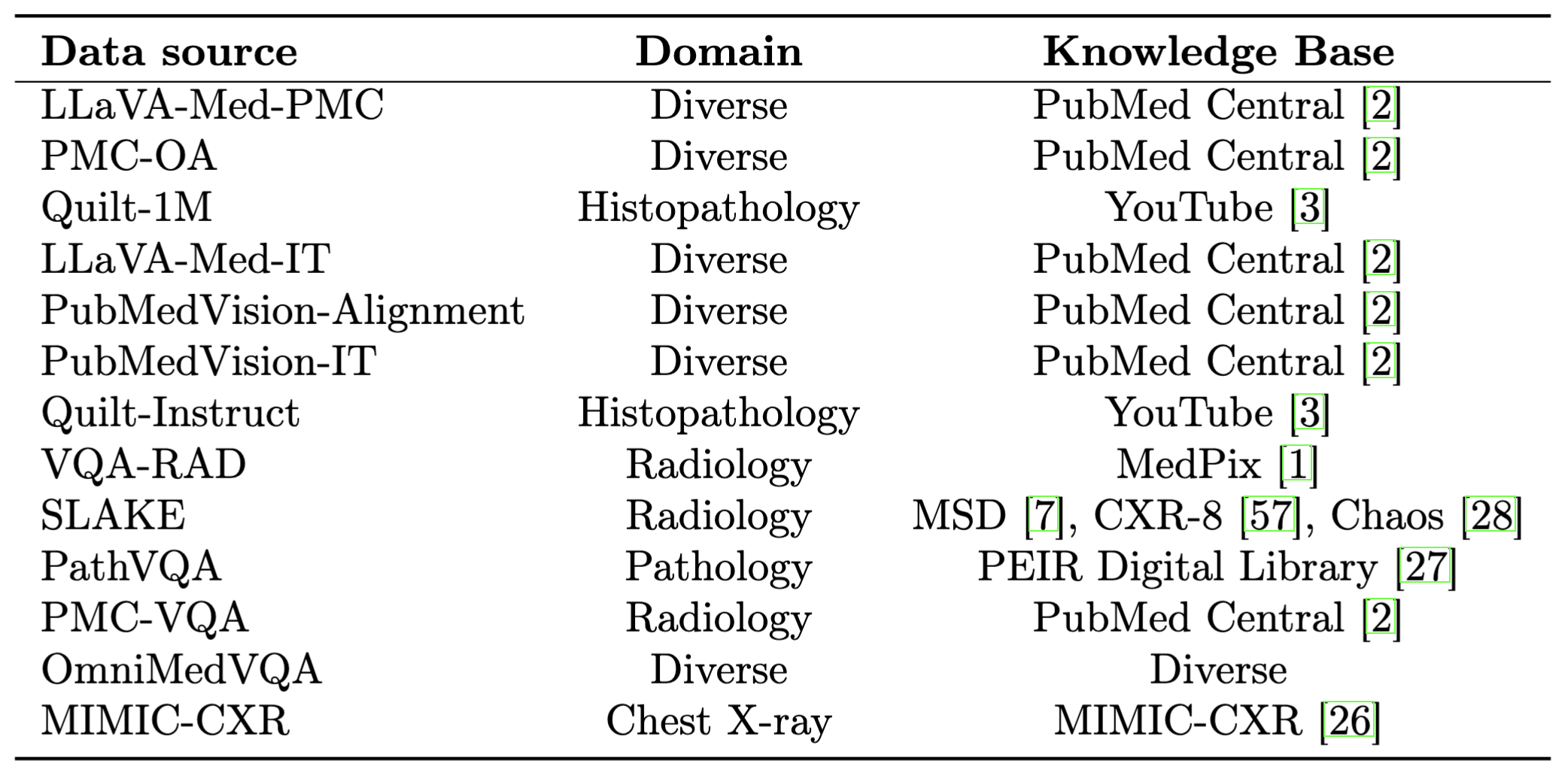
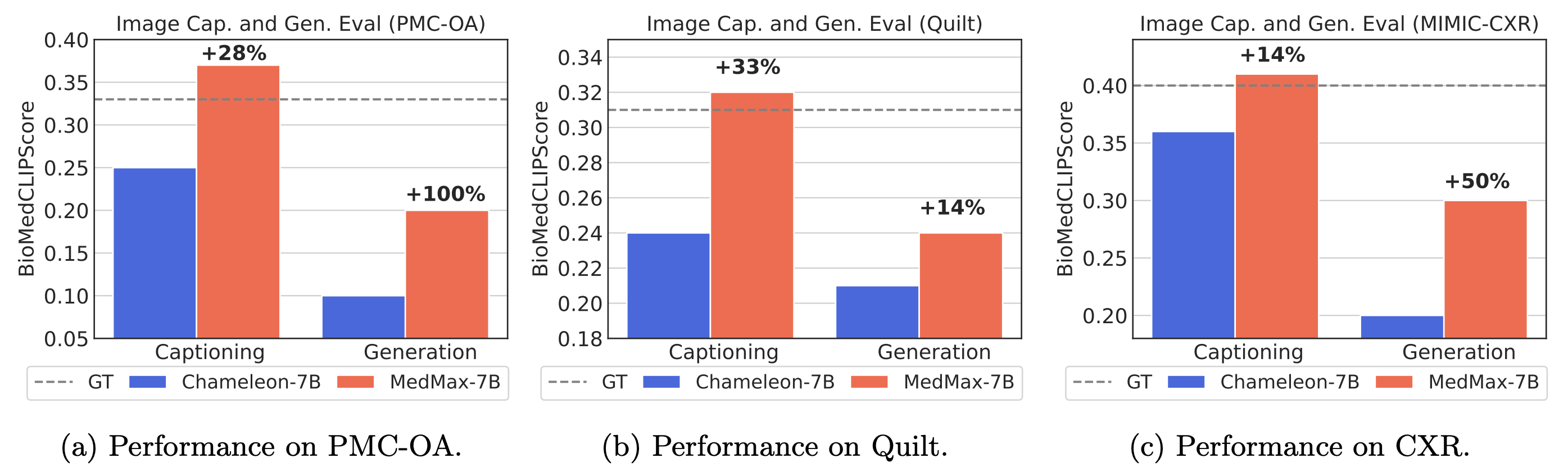
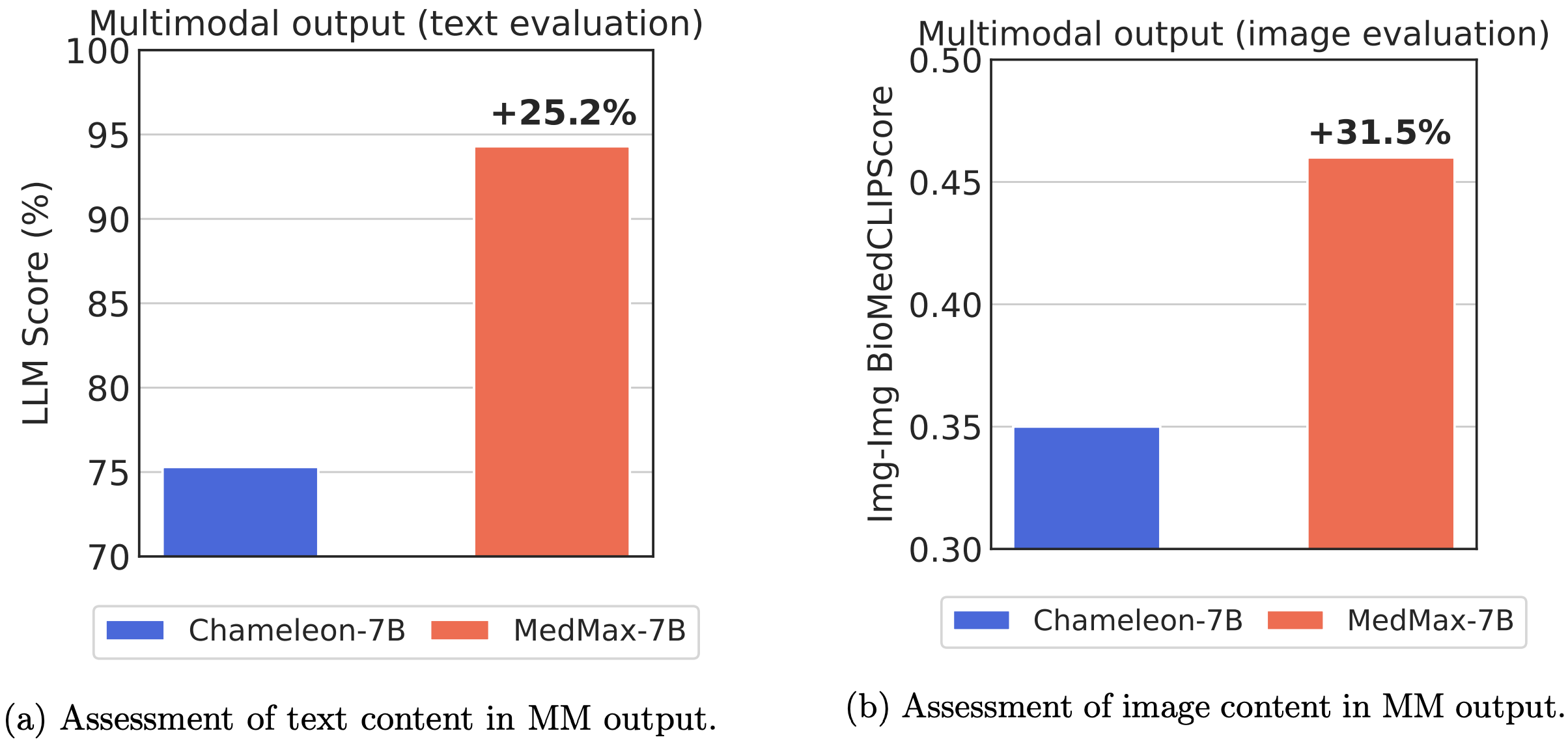
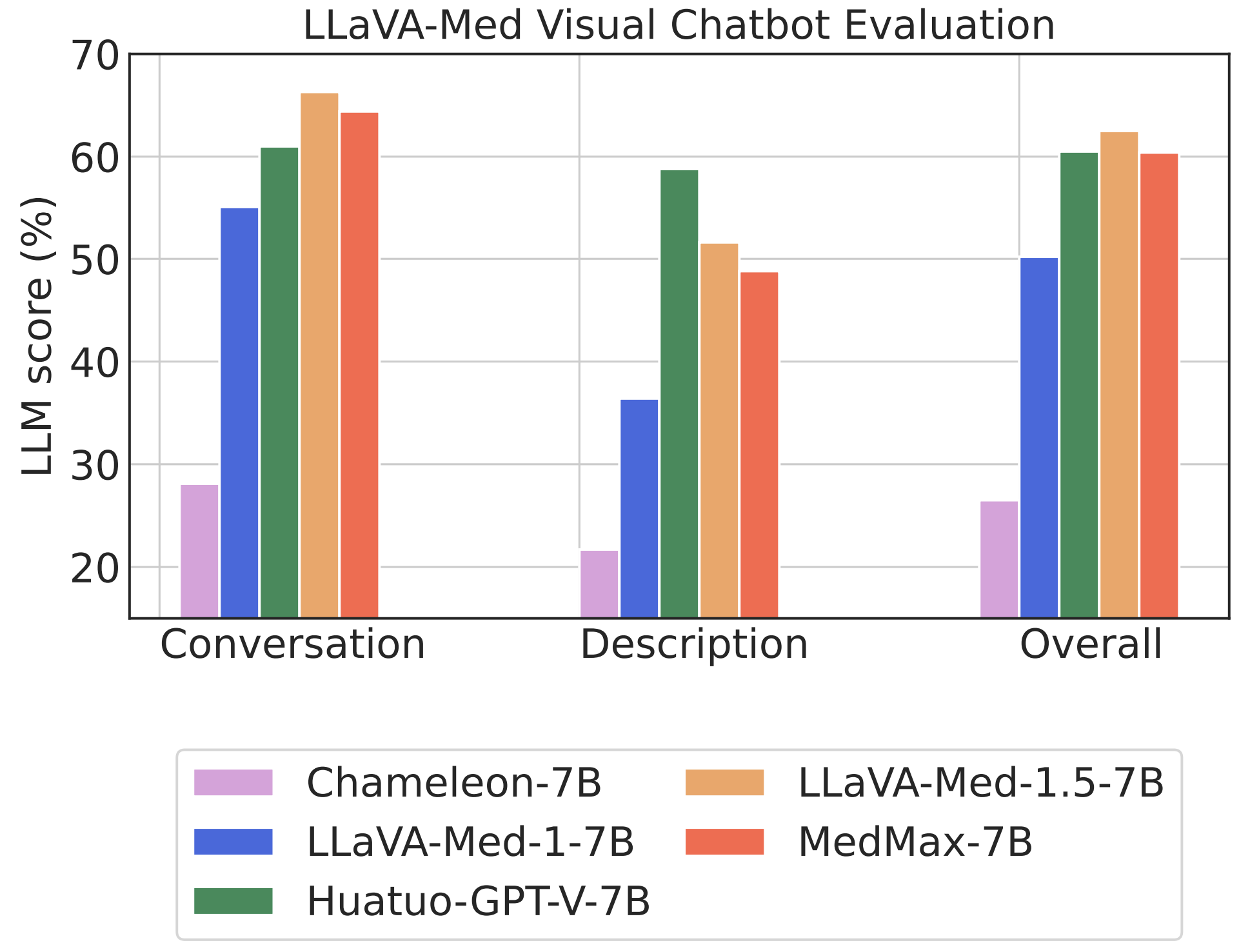
Performance comparison of multimodal models on various biomedical visual question answering datasets.
| Models | Average | VQA-RAD (Closed) | SLAKE (Closed) | PathVQA (Closed) | QuiltVQA (Closed) | VQA-RAD (Open) | SLAKE (Open) | PathVQA (Open) | QuiltVQA (Open) | PMC-VQA | OmniMedVQA | PathMMU | ProbMed |
| Chameleon (7B) | 39.4 | 48.6 | 59.1 | 58.9 | 71.4 | 32.0 | 5.3 | 18.0 | 15.3 | 31.0 | 45.7 | 34.5 | 52.8 |
| LLaVA-Med (v1.5-7B) | 36.6 | 61.0 | 48.7 | 62.7 | 63.0 | 23.0 | 25.1 | 6.2 | 17.2 | 18.9 | 28.7 | 29.8 | 58.5 |
| GPT-4o (mini) | 38.5 | 55.8 | 50.4 | 48.7 | 38.5 | 13.0 | 49.3 | 7.3 | 28.0 | 39.6 | 45.1 | 35.6 | 50.6 |
| GPT-4o | 42.0 | 54.2 | 50.1 | 59.2 | 44.6 | 17.6 | 63.7 | 9.1 | 36.1 | 40.8 | 40.9 | 39.1 | 48.3 |
| HuatuoGPT (Vision-7B) | 52.4 | 74.5 | 70.7 | 65.9 | 55.7 | 19.0 | 53.3 | 6.0 | 22.2 | 51.6 | 75.6 | 55.4 | 78.7 |
 MedMax (7B)
MedMax (7B) |
65.5 | 75.3 | 88.4 | 91.8 | 61.2 | 46.5 | 82.2 | 40.6 | 26.0 | 49.0 | 99.5 | 49.3 | 75.8 |